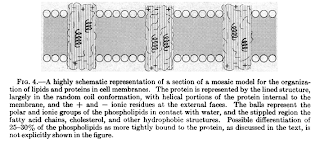
Combining these results (and many others not mentioned), Singer & Nicolson (1972) proposed a model of membrane structure that still holds up. Their model recognized that the main structural component of the membrane (the matrix into which all other components were incorporated) was a lipid bilayer. Most proteins associated with the membrane are embedded within it (Singer & Nicolson called these integral proteins); a smaller fraction of membrane-associate proteins were attached to either the inner or outer surface of the bilayer (referred to as peripheral proteins). The components – phospholipids and proteins – moved around along the surface (either inner or outer) by diffusion. Because proteins embedded in the matrix of lipids created a mosaic of proteins and lipids and the molecules of the membrane moved around each other like molecules of a liquid, Singer & Nicolson referred to their conception of membrane structure as the Fluid Mosaic Model.
Since 1972, specifics of the model have been altered (but I’m not aware of any major changes). In the mid-1970s, Joseph Schlessinger and his colleagues were measuring the diffusion rates of proteins and lipids in the membrane. (Recall that Frye & Edidin already had shown that molecules in the membrane moved around.) Schlessinger and coworkers used a technique known then as fluorescence photobleaching recovery (FRP), but which is now known as fluorescence recovery after photobleaching (FRAP). To use FRAP, fluorescent molecules are attached to other molecules on the surface of the cell – either a particular type of protein that’s common in the cell membrane, for example, or to the phospholipids; you can imagine a cell whose surface is glowing due to the fluorescent molecules. Then, scientists shine a laser at a small spot of the membrane. The energy from the laser excites the fluorescent molecules in the spot exposed to the beam; too much of this will cause the fluorescent molecules to stop fluorescing. They’re bleached, so there is no “glow” in the spot where the laser was aimed. FRAP measures how long it takes for the glow to return to the spot – this provides a measure of how fast unbleached molecules move into the bleached area. One of the interesting results of this work was that when you label proteins with the fluorescent molecules, the fluorescence of the bleached spot never quite returns to previous levels (see graph below). The scientists concluded that this meant not all proteins move – some of the proteins in the bleached area remained there. This wasn’t the case with lipids, so lipids move more freely than (at least some) proteins.
[Figure from Schlessinger and others (1976) PNAS 73(7): 2409-2413; click image for larger view.]
Subsequent research as shown that even membrane proteins that are able to move may still be restricted in some fashion (summarized by Jacobson and his coauthors in 1995). Also, researchers have found that certain types of lipids can aggregate together (along with certain proteins) to form rafts. The components of the raft stay together, but the raft as a unit will move around in the cell membrane (see atomic force micrograph below). Some details of rafts were summarized by Simons & Ikonen in 1997 (a little dated, I know).

[Atomic force micrograph showing membrane rafts “floating” in sea of other lipids. From D.L. Nelson & M.M. Cox (2005) Lehninger’s Principles of Biochemistry, 4th Edition. WH Freeman & Co, p. 385.]
References:
Frye, L.D. and Edidin, M. (1970) “The rapid intermixing of cell surface antigens after formation of mouse-human heterokaryons.” Journal of Cell Science 7: 319-335
Jacobsen, K., Sheets, E.D., and Simson, R. (1995) “Revisiting the Fluid Mosaic Model of membranes.” Science 268: 1441-2